Microscopy: At the heart of images thanks to algorithms
For a long time, Olivia Mariani has been interested in life sciences and research in biology. After a master's degree in image processing, working on microscopy images was the next logical step for her. She looks back with us on her work and years at Idiap.
What did you work on for your thesis?
The subject of my thesis focuses on the development of images of the heart of the zebrafish. It is a widely used animal model because it is, as we say, microscopically small, and transparent. It can also be studied alive and its heart has characteristics very similar to that of a human. Usually, when you use a microscope to study it, you get layered images. There are often deformations called artefacts. In this case, you need a faster microscope, which is also more expensive, otherwise you can use a software that assists the machine. This second method makes it possible to obtain better images and study fundamental biological processes, such as cardiac abnormalities or tissue regeneration. So, as part of my research, I have developed several computational methods to assemble imagery of the heart as it moves and in real time. The goal is for labs that have a scanning microscope available to get better images without having to invest in a faster microscope. It’s more advantageous and less expensive.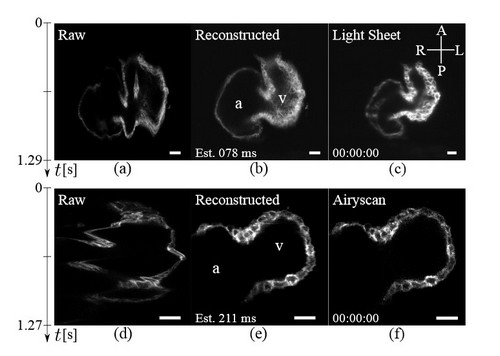
Did you encounter any difficulties while working on such a topic?
Obviously, there were some challenges. Learning the tools first. With digital reconstruction, images are obtained from several sensors and must then be correctly assembled to have a complete image. For the reconstruction of a full sequence of a heartbeat, it’s also necessary to use images acquired over several beats. The most important is to identify good and usable pictures. It’s possible to demonstrate that the reconstruction is successful through simulations and by using a faster microscope to compare images. Fortunately, there is such a microscope at Idiap in the lab of Michael Liebling [editor’s note: head of the Computational bioimaging group and Mariani’s thesis supervisor].
What accomplishment are you most proud of with this thesis?
I am very proud to have participated in the creation of a tool that comes out of the lab. In the beginning, there was no software using these calculation methods to perform digital reconstructions of cardiac imaging and, today, the open source software is available for everybody on Idiap’s website. Of course, there are still some aspects to improve, like the size of the data produced and the accuracy in the faster phases of the heartbeat, but it’s perfectly applicable and works very well for a steady rhythm.
How would you define your years at Idiap?
It was really nice. There are plenty of very qualified people and the latest hardware is available for our research. Technical assistance is also very responsive and efficient. And even if the colleagues I interacted most were the ones in my own office or on the same floor, there are outdoor activities organized by Idiap, which allow you to meet and learn more about those you don't know or don't usually see around.
More information
- Olivia Mariani Thesis – Computational methods for live heart imaging with speed-constrained microscopes
- Olivia Mariani website
- Computational Bioimaging group