VERA FingerVein
Description
VERA FingerVein is a dataset for fingervein recognition and fingervein presentation attack detection (anti-spoofing). The dataset consists of 440 NIR Bona-Fide images from 110 clients captured using an Open Sensor. The dataset also contains presentation attacks (spoofing attacks) to the same 440 images that can be used to study vulnerability of vein recognition systems or to develop presentation attack detection techniques.
Reference
If you use this database in your publication, please cite the following publication:
Pedro Tome, Ramachandra Raghavendra, Christoph Busch, Santosh Tirunagari, Norman Poh, B. H. Shekar, Diego Gragnaniello, Carlo Sansone, Luisa Verdoliva and Sébastien Marcel : "The 1st Competition on Counter Measures to Finger Vein Spoofing Attacks", in Proceedings of The 8th IAPR International Conference on Biometrics (ICB), 2015
10.1109/ICB.2015.7139067
http://publications.idiap.ch/index.php/publications/show/3095
Data
All fingervein samples have been recorded using the open finger vein sensor described in [BT12]. A total of 110 subjects presented their 2 indexes to the sensor in a single session and recorded 2 samples per finger with 5 minutes separation between the 2 trials. The database, therefore, contains a total of 440 samples and 220 unique fingers.
The recordings were performed at 2 different locations, always inside buildings with normal light conditions. The data for the first 78 subjects derives from the the first location while the remaining 32 come from the second location.
The dataset is composed of 40 women and 70 men whose ages are between 18 and 60 with an average at 33. Information about gender and age of subjects are provided with our dataset interface.
Samples are stored as follow with the following filename convention: full/bf/004-F/004_L_2. The fields can be interpreted as <size>/<source>/<subject-id>-<gender>/<subject-id>_<side>_<trial>. The <size> represents one of two options full or cropped. The images in the full directory contain the full image produced by the sensor. The images in the cropped directory represent pre-cropped region-of-interests (RoI) which can be directly used for feature extraction without region-of-interest detection. We provide both verification and presentation-attack detection protocols for full or cropped versions of the images.
Note
Images in the cropped subdirectory where simply derived from images in the full directory by cropping a fixed amount of 50 pixels from each border of the input image.
The <source> field may one of bf (bona fide) or pa (presentation attack) and represent the genuiness of the image. Naturally, biometric recognition uses only images of the bf folder for all protocols as indicated below. The <subject-id> is a 3 digits number that stands for the subject's unique identifier. The <gender> value can be either M (male) or F (female). The <side> corresponds to the index finger side and can be set to either "R" or "L" ("Right" or "Left"). The <trial> corresponds to either the first (1) or the second (2) time the subject interacted with the device.
Images in the full folder are stored in PNG format, with a size of 250x665 pixels (height, width). Size of the files is around 80 kbytes per sample.
Here are examples of bona fide (bf) samples from the full folder inside the database, for subject 029-M:

Image from subject 0029 (male). This image corresponds to the first trial for the left index finger.
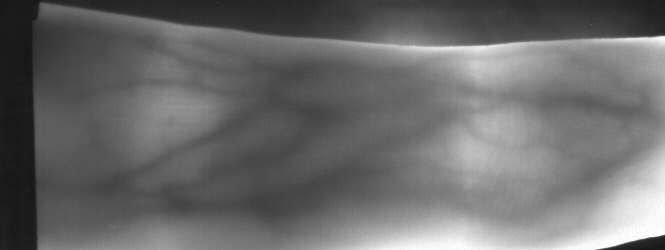
Image from subject 0029 (male). This image corresponds to the second trial for the left index finger.
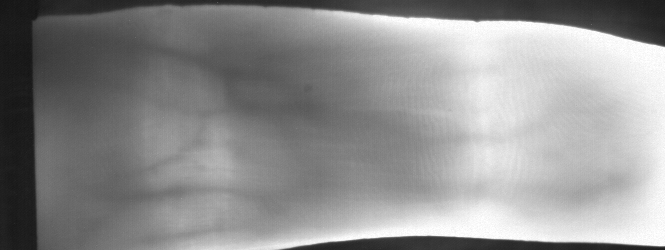
Image from subject 0029 (male). This image corresponds to the first trial for the right index finger.
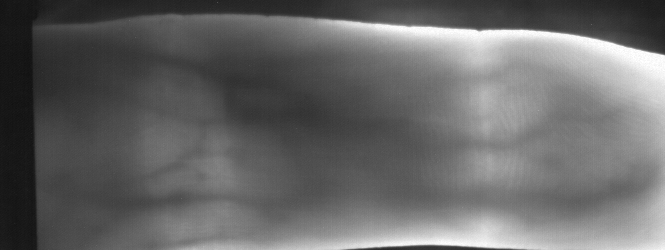
Image from subject 0029 (male). This image corresponds to the second trial for the right index finger.
Images in the cropped folder are stored in PNG format, with a size of 150x565 pixels (height, width). Because of the simplified sensor design and fixed finger positioning, cropping was performed by simply removing 50 pixels from each border of the original raw image. Size of the files is around 40 kbytes per sample.
Here are examples of bona fide (bf) samples from the cropped folder inside the database, for subject 029-M:

Image from subject 0029 (male). This image corresponds to the first trial for the left index finger. This version contains only the pre-cropped region-of-interest.

Image from subject 0029 (male). This image corresponds to the second trial for the left index finger. This version contains only the pre-cropped region-of-interest.

Image from subject 0029 (male). This image corresponds to the first trial for the right index finger. This version contains only the pre-cropped region-of-interest.

Image from subject 0029 (male). This image corresponds to the second trial for the right index finger. This version contains only the pre-cropped region-of-interest.
Biometric Data Acquisition Protocol
Subjects were asked to put their index in the sensor and then adjust the position such that the finger is on the center of the image. Bram Ton's Graphical User Interface (GUI) was used for visual feedback, Near Infra Red light control and acquisition. When the automated light control was performing unproperly the operator adjusted manually the intensities of the leds to achieve a better contrast of the vein pattern.
Subjects first presented an index, then the other, a second time the first index and a second time the second index. The whole process took around 5 minutes per subject in average.
The file metadata.csv contains additional information of gender and age (at the time of capture) for each of the 110 individuals available in the dataset.
Presentation-Attack Protocol
To create effective presentation attacks for this dataset, images available were printed on high-quality (200 grams-per-square-meter - GSM) white paper using a laser printer (toner can absorb to near-infrared light used in fingervein sensors), and presented to the same sensor. More information and details can be found on Section 2.2 of the original publication [TVM14].
All presentation attacks were recorded using the same open finger vein sensor used to record the Biometric Recognition counterpart. Images are stored in PNG format, with a size of 250x665 pixels (height, width). Files are named in a matching convention to their counterparts in the biometric recognition. Size of the files is around 80 kbytes per sample.
All bonafide samples corresponds to unaltered originals from the bf part of the dataset.
Images in the full folder are stored in PNG format, with a size of 250x665 pixels (height, width). Size of the files is around 80 kbytes per sample.
Here are examples of presentation-attack (pa) samples from the full folder inside the database, for subject 029-M:
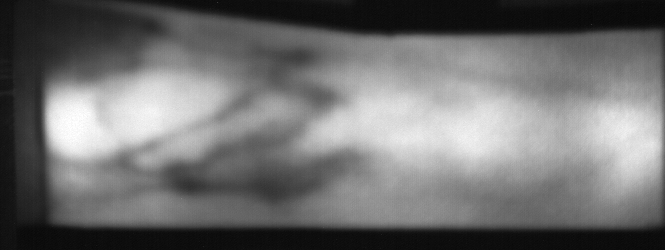
Image from subject 0029 (male). This image corresponds to the first trial for the left index finger.
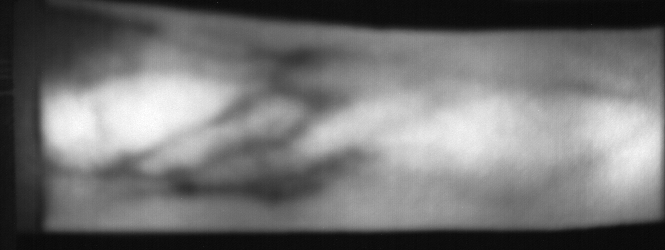
Image from subject 0029 (male). This image corresponds to the second trial for the left index finger.
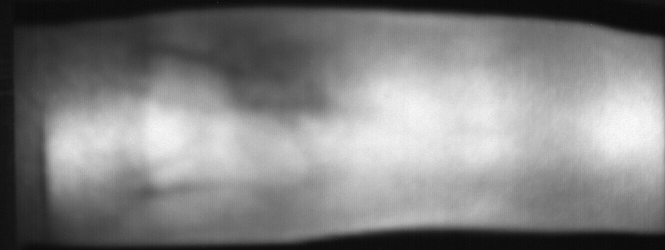
Image from subject 0029 (male). This image corresponds to the first trial for the right index finger.
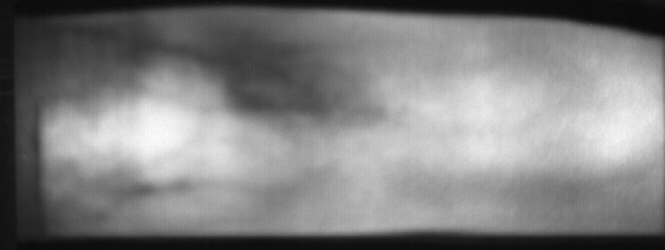
Image from subject 0029 (male). This image corresponds to the second trial for the right index finger.
Images in the cropped folder are stored in PNG format, with a size of 150x565 pixels (height, width). Cropping happened in the same way as for the original biometric recognition subset. The size of the files is around 40 kbytes per sample.
Here are examples of presentation-attack (pa) samples from the cropped folder inside the database, for subject 029-M:

Image from subject 0029 (male). This image corresponds to the first trial for the left index finger. This version contains only the pre-cropped region-of-interest.

Image from subject 0029 (male). This image corresponds to the second trial for the left index finger. This version contains only the pre-cropped region-of-interest.

Image from subject 0029 (male). This image corresponds to the first trial for the right index finger. This version contains only the pre-cropped region-of-interest.

Image from subject 0029 (male). This image corresponds to the second trial for the right index finger. This version contains only the pre-cropped region-of-interest.
Region-of-Interest Annotations
This repository contains the annotations for the fingervein recognition "VERA" fingervein dataset. Each annotation is a text file with points which mark the region-of-interest (RoI) in each image, containing the finger region and excluding the background. To make use of the annotations, you must join the points creating a polygon.
Each annotation file contains annotation for a single, matching image in the original raw dataset. Each file is composed of as many lines as points annotated. There isn't a fixed number of annotations per file. The number of annoated points depends only on the finger contour - some fingers will have therefore more annotations than others. Each point is represented as two (16-bit) unsigned integer numbers representing the y and x coordinates in this order. Here is an example Python code using numpy that can read the annotations and return a 2D-array with them:
import numpy
return numpy.loadtxt('/path/to/annotation/file.txt', dtype='uint16')
Annotations cover only raw data in the full dataset directory. We don't provide annotations for the data in the cropped directory for obvious reasons.
Protocols
You'll find a directory called protocols which is distributed with the dataset. This directory contains filelists for each protocol that has even been published with the above-cited publications. Protocols are divided into two categories: biometric recognition (and vulnerability analysis), in the bio sub-directory and presentation attack detection, in the pad sub-directory. There are 4 protocols for biometric recognition and 2 protocols for presentation attack detection. Each biometric recognition protocol can also be used in two variants: cropped (using pre-cropped regions-of-interest) and va (for vulnerability analysis). They are described next.
The "bio/Nom" protocol
The "Nom" (normal operation mode) protocol corresponds to the standard biometric verification scenario. For the VERA database, each finger for all subjects will be used for enrolling and probing. Data from the first trial is used for enrollment, while data from the second trial is used for probing. Matching happens exhaustively. In summary:
- 110 subjects x 2 fingers = 220 unique fingers
- 2 trials per finger, so 440 unique images
- Use trial 1 for enrollment and trial 2 for probing
- Total of 220 genuine scores and 220x219 = 48180 impostor scores
- No images for training
Note
The cropped variant of this protocol simply uses images in the cropped directory instead of those in the full directory. For vulnerability analysis, use either images in the full directory or cropped directory, but match also against probes in the */pa directory respecting the probing part of this protocol.
The "bio/Fifty" protocol
The "Fifty" protocol is meant as a reduced version of the "Nom" protocol, for quick check purposes. All definitions are the same, except we only use the first 50 subjects in the dataset (numbered 1 until 59). In summary:
- 50 subjects x 2 fingers = 100 unique fingers
- 2 sessions per finger, so 200 unique images
- Use trial sample 1 for enrollment and trial sample 2 for probing
- Total of 100 genuine scores and 100x99 = 9900 impostor scores
- Use all remaining images for training (440-200 = 240 images). In this case, the remaining images all belong to different subjects that those on the development set.
Note
The cropped variant of this protocol simply uses images in the cropped directory instead of those in the full directory. For vulnerability analysis, use either images in the full directory or cropped directory, but match also against probes in the */pa directory respecting the probing part of this protocol.
The "bio/B" protocol
The "B" protocol was created to simulate a biometric recognition evaluation scenario similar to that from the UTFVP database (see [BT12]). 108 unique fingers were picked:
- Each of the 2 fingers from the first 48 subjects (96 unique fingers), subjects numbered from 1 until 57
- The left fingers from the next 6 subjects (6 unique fingers), subjects numbered from 58 until 65
- The right fingers from the next 6 subjects (6 unique fingers), subjects numbered from 66 until 72
Then, protocol "B" was setup in this way:
- 108 unique fingers
- 2 trials per finger, so 216 unique images
- Match all fingers against all images (even against itself)
- Total of 216x2 = 432 genuine scores and 216x214 = 46224 impostor scores
- Use all remaining images for training (440-216 = 224 samples). In this case, the remaining images not all belong to different subjects that those on the development set.
Note
The cropped variant of this protocol simply uses images in the cropped directory instead of those in the full directory. For vulnerability analysis, use either images in the full directory or cropped directory, but match also against probes in the */pa directory respecting the probing part of this protocol.
The "bio/Full" protocol
The "Full" protocol is similar to protocol "B" in the sense it tries to match all existing images against all others (including itself), but uses all subjects and samples instead of a limited set. It was conceived to facilitate cross-folding tests on the database. So:
- 220 unique fingers
- 2 trials per finger, so 440 unique images
- Match all fingers against all images (even against itself)
- Total of 440x2 = 880 genuine scores and 440x438 = 192720 impostor scores
- No samples are available for training in this protocol
Note
The cropped variant of this protocol simply uses images in the cropped directory instead of those in the full directory. For vulnerability analysis, use either images in the full directory or cropped directory, but match also against probes in the */pa directory respecting the probing part of this protocol.
The "pad/full" Protocol
This is a presentation attack detection protocol. It allows for training and evaluating binary-decision making counter-measures to Presentation Attacks. The available data comprised of bonafide and presentation attacks are split into 3 sub-groups:
- Training data ("train"), to be used for training your detector;
- Development data ("dev"), to be used for threshold estimation and fine-tunning;
- Test data ("test"), with which to report error figures;
Clients that appear in one of the data sets (train, devel or test) do not appear in any other set.
In this protocol, the full image as captured from the sensor is available to the user. Here is a summary:
- Training set: 30 subjects (identifiers from 1 to 31 inclusive). There are 240 samples on this set.
- Development set: 30 subjects (identifiers from 32 to 72 inclusive). There are 240 samples on this set.
- Test set: 50 subjects (identifiers from 73 to 124 inclusive). There are 400 samples on this set.
The "pad/cropped" Protocol
In this protocol, only a pre-cropped image of size 150x565 pixels (height, width) is provided to the user, that can skip region-of-interest detection on the processing toolchain. The objective is to test user algorithms don't rely on information outside of the finger area for presentation attack detection. The subject separation is the same as for protocol "full".
Canonical Implementation
We provide a canonical iterator implementation allowing quick programmatic access to the raw data provided by this dataset respecting the protocols above. You'll find this implementation in the package bob.db.verafinger, which is part of the Bob framework. Please address questions regarding this software to our mailing list.
| [BT12] | (1, 2) B. Ton. Vascular pattern of the finger: biometric of the future? Sensor design, data collection and performance verification. Masters Thesis, University of Twente, Netherlands, July 2012. |
| [TVM14] | Pedro Tome, Matthias Vanoni and Sébastien Marcel, On the Vulnerability of Finger Vein Recognition to Spoofing, in: IEEE International Conference of the Biometrics Special Interest Group (BIOSIG), Darmstadt, Germay, pages 1 - 10, IEEE, 2014 |