Dask: Scale your scikit.learn pipelines¶
Dask is a flexible library for parallel computing in Python. The purpose of this guide is not to describe how dask works. For that, go to its documentation. Moreover, there are plenty of tutorials online. For instance, this official one; a nice overview was presented in AnacondaCon 2018 and there’s even one crafted for Idiap.
The purpose of this guide is to describe:
The integration of dask with scikit learn pipelines and samples
The specificities of Dask under the Idiap SGE
From Scikit Learn pipelines to Dask Task Graphs¶
The purpose of scikit learn pipelines is to assemble several scikit estimators in one final one. Then, it is possible to use the methods fit and transform to create models and transform your data respectivelly.
Any pipeline can be transformed in a Dask Graph to be further executed by any Dask Client.
This is carried out via the wrap function when used like wrap(["dask"], estimator) (see Convenience wrapper function).
Such function does two things:
Edit the current
sklearn.pipeline.Pipelineby adding a new first step, where input samples are transformed in Dask Bag. This allows the usage ofdask.bag.mapfor further transformations.Wrap all estimators in the pipeline with
DaskWrapper. This wrapper is responsible for the creation of the task graph for the methods .fit and .transform.
The code snippet below enables such feature for an arbitrary pipeline.
>>> import bob.pipelines as mario
>>> from sklearn.pipeline import make_pipeline
>>> pipeline = make_pipeline(...)
>>> dask_pipeline = mario.wrap(["dask"], pipeline) # Create a dask graph
>>> dask_pipeline.fit_transform(....).compute() # Run the task graph using the default client
The code below is an example. Especially lines 59-63 where we convert such pipeline in a Dask Graph and runs it in a local computer.
1import os
2import shutil
3
4import numpy
5
6from sklearn.base import BaseEstimator, TransformerMixin
7from sklearn.pipeline import make_pipeline
8
9import bob.pipelines
10
11from bob.pipelines.sample import Sample
12
13
14class MyTransformer(TransformerMixin, BaseEstimator):
15 def transform(self, X, metadata=None):
16 # Transform `X` with metadata
17 return X
18
19 def fit(self, X, y=None):
20 pass
21
22 def _more_tags(self):
23 return {"requires_fit": False}
24
25
26class MyFitTranformer(TransformerMixin, BaseEstimator):
27 def __init__(self):
28 self._fit_model = None
29
30 def transform(self, X, metadata=None):
31 # Transform `X`
32 return [x @ self._fit_model for x in X]
33
34 def fit(self, X):
35 self._fit_model = numpy.array([[1, 2], [3, 4]])
36 return self
37
38
39# Creating X
40X = numpy.zeros((2, 2))
41# Wrapping X with Samples
42X_as_sample = [Sample(X, key=str(i), metadata=1) for i in range(10)]
43
44# Building an arbitrary pipeline
45model_path = "./dask_tmp"
46os.makedirs(model_path, exist_ok=True)
47pipeline = make_pipeline(MyTransformer(), MyFitTranformer())
48
49# Wrapping with sample, checkpoint and dask
50pipeline = bob.pipelines.wrap(
51 ["sample", "checkpoint", "dask"],
52 pipeline,
53 model_path=os.path.join(model_path, "model.pickle"),
54 features_dir=model_path,
55 transform_extra_arguments=(("metadata", "metadata"),),
56)
57
58# Create a dask graph from a pipeline
59# Run the task graph in the local computer in a single tread
60X_transformed = pipeline.fit_transform(X_as_sample).compute(
61 scheduler="single-threaded"
62)
63
64
65shutil.rmtree(model_path)
Such code generates the following graph.
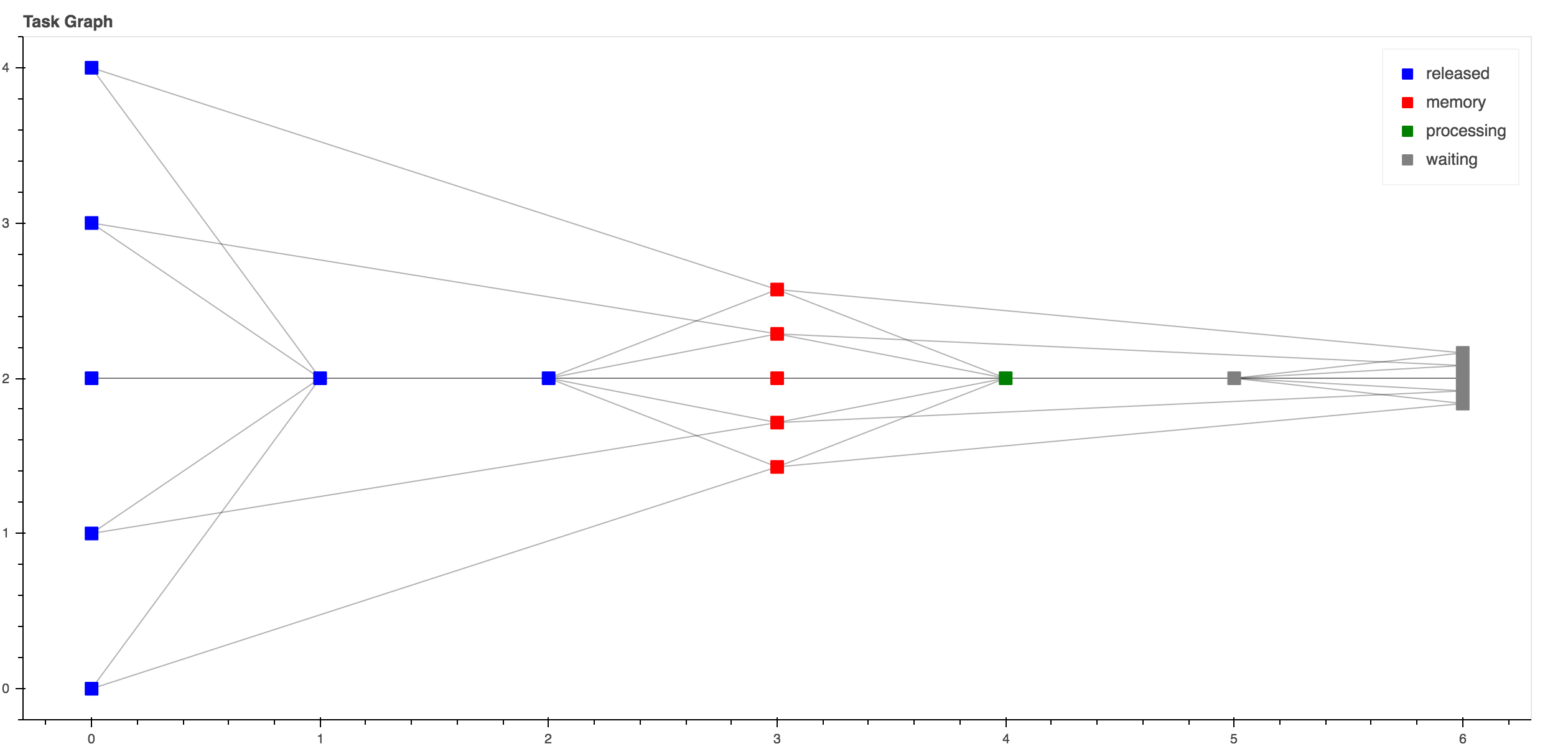
Fig. 22 This graph can be seem by running http://localhost:8787 during its execution.¶
Dask + Idiap SGE¶
Dask, allows the deployment and parallelization of graphs either locally or in complex job queuing systems, such as PBS, SGE…. This is achieved via Dask-Jobqueue. Below follow a nice video explaining what is the Dask-Jobqueue, some of its features and how to use it to run dask graphs.
Warning
To submit jobs at Idiap’s SGE it’s mandatory to set a project name. Run the code below to set it:
$ bob config set sge.project <YOUR_PROJECT_NAME>
The snippet below shows how to deploy the exact same pipeline from the previous section in the Idiap SGE cluster
>>> from bob.pipelines.distributed.sge import SGEMultipleQueuesCluster
>>> from dask.distributed import Client
>>> cluster = SGEMultipleQueuesCluster() # Creates the SGE launcher that launches jobs in the q_1day
>>> client = Client(cluster) # Creates the scheduler and attaching it to the SGE job queue system
>>> dask_pipeline.fit_transform(....).compute(scheduler=client) # Runs my graph in the Idiap SGE
That’s it, you just run a scikit pipeline in the Idiap SGE grid :-)
Dask provides generic deployment mechanism for SGE systems, but it contains the following limitations:
It assumes that a dask graph runs in an homogeneous grid setup. For instance, if parts your graph needs a specific resource that it’s avaible in other SGE queues (e.g q_gpu, q_long_gpu, IO_BIG), the scheduler is not able to request those resources on the fly.
As a result of 1., the mechanism of adaptive deployment is not able to handle job submissions of two or more queues.
For this reason the generic SGE laucher was extended to this one bob.pipelines.distributed.sge.SGEMultipleQueuesCluster. Next subsections presents some code samples using this launcher in the most common cases you will probably find in your daily job.
Launching jobs in different SGE queues¶
SGE queue specs are defined in python dictionary as in the example below, where, the root keys are the labels of the SGE queue and the other inner keys represents:
queue: The real name of the SGE queue
memory: The amount of memory required for the job
io_big: Submit jobs with IO_BIG=TRUE
resource_spec: Whatever other key using in qsub -l
resources: Reference label used to tag dask delayed so it will run in a specific queue. This is a very important feature the will be discussed in the next section.
>>> Q_1DAY_GPU_SPEC = {
... "default": {
... "queue": "q_1day",
... "memory": "8GB",
... "io_big": True,
... "resource_spec": "",
... "max_jobs": 48,
... "resources": "",
... },
... "q_short_gpu": {
... "queue": "q_short_gpu",
... "memory": "12GB",
... "io_big": False,
... "resource_spec": "",
... "max_jobs": 48,
... "resources": {"q_short_gpu":1},
... },
... }
Now that the queue specifications are set, let’s trigger some jobs.
>>> from bob.pipelines.distributed.sge import SGEMultipleQueuesCluster
>>> from dask.distributed import Client
>>> cluster = SGEMultipleQueuesCluster(sge_job_spec=Q_1DAY_GPU_SPEC)
>>> client = Client(cluster) # Creating the scheduler
Note
To check if the jobs were actually submitted always do qstat:
$ qstat
Running estimator operations in specific SGE queues¶
Sometimes it’s necessary to run parts of a pipeline in specific SGE queues (e.g. q_1day IO_BIG or q_gpu). The example below shows how this is approached (lines 52 to 57). In this example, the fit method of MyBoostedFitTransformer runs on q_short_gpu
1import os
2import shutil
3
4import numpy
5
6from dask.distributed import Client
7from sklearn.base import BaseEstimator, TransformerMixin
8from sklearn.pipeline import make_pipeline
9
10import bob.pipelines
11
12from bob.pipelines.distributed.sge import (
13 SGEMultipleQueuesCluster,
14 get_resource_requirements,
15)
16from bob.pipelines.sample import Sample
17
18
19class MyTransformer(TransformerMixin, BaseEstimator):
20 def transform(self, X, metadata=None):
21 # Transform `X` with metadata
22 return X
23
24 def fit(self, X, y=None):
25 pass
26
27 def _more_tags(self):
28 return {"requires_fit": False}
29
30
31class MyFitTranformer(TransformerMixin, BaseEstimator):
32 def __init__(self):
33 self._fit_model = None
34
35 def transform(self, X, metadata=None):
36 # Transform `X`
37 return [x @ self._fit_model for x in X]
38
39 def fit(self, X):
40 self._fit_model = numpy.array([[1, 2], [3, 4]])
41 return self
42
43
44# Creating X
45X = numpy.zeros((2, 2))
46# Wrapping X with Samples
47X_as_sample = [Sample(X, key=str(i), metadata=1) for i in range(10)]
48
49# Building an arbitrary pipeline
50model_path = "./dask_tmp"
51os.makedirs(model_path, exist_ok=True)
52pipeline = make_pipeline(MyTransformer(), MyFitTranformer())
53
54# Wrapping with sample, checkpoint and dask
55# NOTE that pipeline.fit will run in `q_short_gpu`
56pipeline = bob.pipelines.wrap(
57 ["sample", "checkpoint", "dask"],
58 pipeline,
59 model_path=model_path,
60 transform_extra_arguments=(("metadata", "metadata"),),
61 fit_tag="q_short_gpu",
62)
63
64# Creating my cluster obj.
65cluster = SGEMultipleQueuesCluster()
66client = Client(cluster) # Creating the scheduler
67resources = get_resource_requirements(pipeline)
68
69# Run the task graph in the local computer in a single tread
70# NOTE THAT resources is set in .compute
71X_transformed = pipeline.fit_transform(X_as_sample).compute(
72 scheduler=client, resources=resources
73)
74
75shutil.rmtree(model_path)